Water quality monitoring of bacteria and viruses using combined concentration and molecular analysis techniques
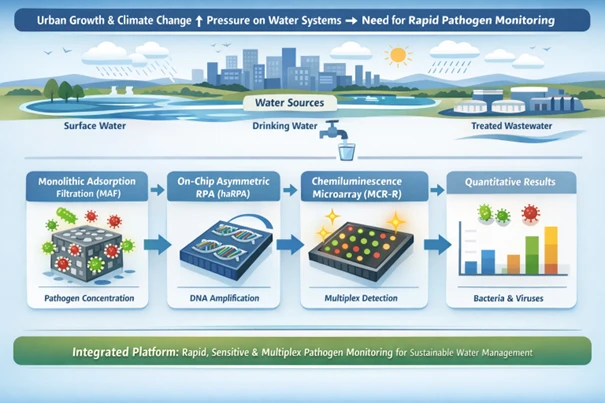
Rapid urban growth and climate change are placing rising demands on urban water resources, leading to increased pollution and pressure on wastewater systems. To address these challenges, an integrated management is essential to provide sustainable and safe water supply. The overall goal of the project UrbaQuantum is to develop on one hand novel sensors developments for the quantification of pathogens, cyanobacteria, microplastic and other contaminants and on the other hand, optimized sensing strategies and signal processing algorithms for cost-effective continuous monitoring of related pollution events. The part of our work package is to develop a multiplex sensing platform for quantitative detection of selected bacteria and viruses from various water sources, including treated wastewater, drinking water, and surface water. The system combines automatic preconcentration of pathogens using monolithic adsorption filtration (MAF) with on-chip molecular analysis via heterogeneous asymmetric recombinase polymerase amplification (haRPA) on a chemiluminescence microarray analysis platform. Epoxy-functionalized macroporous monoliths are optimized for high filtration efficiency, while molecular biology protocols, including primer design, nucleic acid extraction, and amplification, are adapted for microarray-based detection. This integrated system allows for the simultaneous enrichment, amplification, and quantitative detection of targeted pathogens directly on a microchip, enabling rapid and sensitive monitoring. The development of this platform contributes to the safety and sustainability of urban water infrastructures under the UrbaQuantum framework.
References
[1] Hess, S., Niessner, R., & Seidel, M. (2021). Quantitative detection of human adenovirus from river water by monolithic adsorption filtration and quantitative PCR. Journal of Virological Methods, 292, 114128. https://doi.org/10.1016/j.jviromet.2021.114128
[2] Kober, C., Niessner, R., & Seidel, M. (2018). Quantification of viable and non-viable Legionella spp. by heterogeneous asymmetric recombinase polymerase amplification (haRPA) on a flow-based chemiluminescence microarray. Biosensors and Bioelectronics, 100, 49–55. http://dx.doi.org/10.1016/j.bios.2017.08.053
[3] Kunze, A., Dilcher, M., Abd El Wahed, A., Hufert, F., Niessner, R., & Seidel, M. (2016). On-chip isothermal nucleic acid amplification on flow-based chemiluminescence microarray analysis platform for the detection of viruses and bacteria. Analytical Chemistry, 88, 898–905.
http://dx.doi.org/10.1016/j.jviromet.2015.06.007
[4] Pei, L., Rieger, M., Lengger, S., Ott, S., Zawadsky, C., Hartmann, N. M., Selinka, H. C., Tiehm, A., Niessner, R., & Seidel, M. (2012). Combination of crossflow ultrafiltration, monolithic affinity filtration, and quantitative reverse transcriptase PCR for rapid concentration and quantification of model viruses in water. Environmental Science & Technology, 46, 10073–10080. https://doi.org/10.1021/es302304t
Funding Cooperation
EU-funded, Grant agreement ID: 101180452 CyRIC
HORIZON-CL6-2024-ZEROPOLLUTION-02