Exploring Reaction Paths for Sulfonamide Hydrolysis: Insight from Experiments and Computations
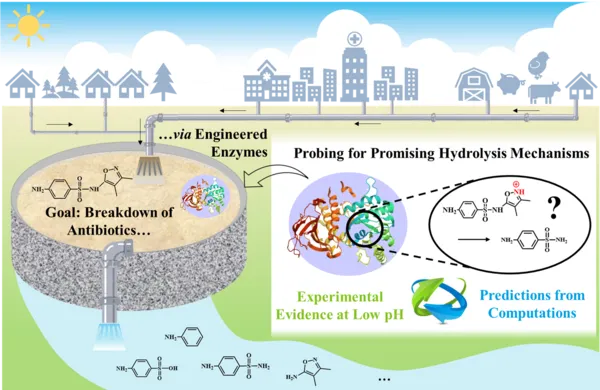
A synergy of experiments and computations is explored to decipher possible hydrolysis mechanisms of sulfonamides. This insight is key to creating novel enzymes with specific functions to remove such antibiotics from water.
Sulfonamides (SAs) have been widely used as antibiotics in human and veterinary medicine. In the environment, they have been detected both in unchanged and in metabolized form as a result of incomplete biotransformation. Antibiotics have attracted particular attention because of their potential to give rise to bacterial antibiotic resistance. Much interest is, therefore, directed at their targeted elimination. While many studies have focused on detecting oxidative biodegradation through associated metabolite identification, only limited information is available on the possibility to break them down via hydrolysis. Therefore, our project probes for putative hydrolysis mechanisms of SAs based on experimental and computational evidence.
Sulphamethoxazole, sulphisoxazole, sulphamethiazole, sulphathiazole, sulphapyridine, sulphadiazine, sulphadimidine and sulphadimethoxine were exposed to a range of pH conditions (2.0, 4.0, 6.0, 8.0, or 10.0) over three months. While most selected SAs were hydrolytically stable in buffer solutions at pH 4.0, 6.0, 8.0, and 10.0, five out of eight SAs (sulphisoxazole, sulphapyridine, sulphadiazine, sulphadimidine, and sulphadimethoxine) were degraded effectively at pH 2.0. In addition, hydrolytic metabolites, such as sulphanilic acid, sulphanilamide, aniline and some of the corresponding leaving groups were identified and quantified by HPLC-UV. We aim to identify further SA transformation products by analysis with Orbitrap high-resolution mass spectrometry (HRMS). In a complementary approach, isotope effects (e.g., 13C/12C, 15N/14N) in individual sulfonamide compounds are measured by gas chromatography - isotope ratio mass spectrometry (GC-IRMS). In combination with quantum chemical calculations, we aim to use this evidence to derive a mechanistic understanding as computational guidance for directed evolution of novel enzymes for curbing micropollutant contamination in drinking and waste water.