Development of Bioorthogonal Noncanonical Amino Acid Tagging – Surface-enhanced Raman Scattering (BONCAT-SERS) to Visualize Active Bacteria Responsible for Degradation of Organic Pollutants
Objectives
- Development the coupling of BONCAT-SERS
- Visualization and characterization of active bacteria-micropollutant degraders
Methods of Approach
- Raman microspectroscopy (RM)
- Surface-enhanced Raman scattering (SERS)
- Fluorescence microscopy
Description
Elimination of organic pollutants is crucial in the production of clean drinking water and for ensuring a sustainable water environment. Such micropollutants as pesticides, pharmaceuticals, personal care products or industrial compounds have received increased attention, because of their frequent detection in waste waters at low concentrations and unknown effects on aquatic ecosystem health. Conventional activated sludge systems employed in waste water treatment plants are typically not designed to completely remove micropollutants. Natural and engineered systems that enhance biotransformation of pollutants are attractive as means of environmentally-friendly and cost-efficient purification. However, it is difficult to identify active cells responsible for the transformation of trace organic chemicals within bacterial community. Mostly, currently available techniques do not disclose who is really metabolically active and growing (gene sequencing, scanning electron and fluorescence microscopy), are expensive and may disturb the studied systems (stable isotope probing) or reveal active cells in time-consuming and cost-intense mode with restriction to explore only cultured microorganisms (immunofluorescence analysis). Thus, a minimally invasive, rapid, and sensitive approach that combines visualization and characterization of active cells is warranted.
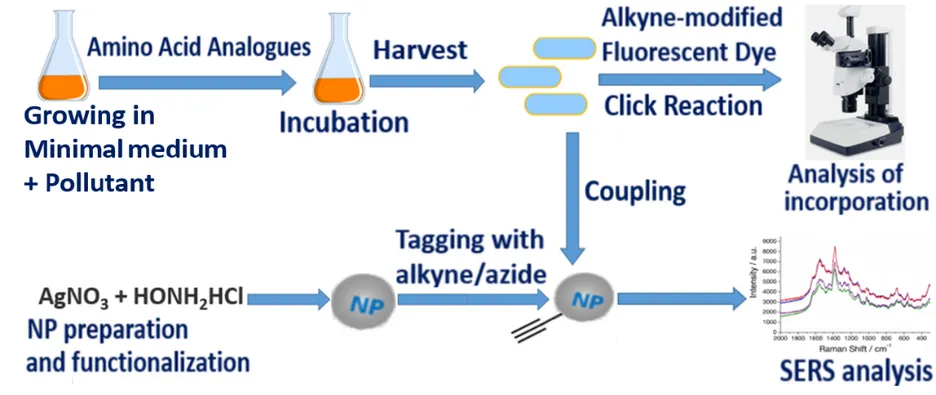
Visualization of active cells can be catalyzed by bioorthogonal noncanonical amino acid tagging (BONCAT). This emerging method is used to study individual cell response to external signals in situ and relies on the in vivo incorporation of amino acids surrogates into bacterial biomass. Amino acid analogues (AAAs) with modifiable tags are included into polypeptide chains by metabolically active cells. Active bacteria can subsequently be visualized by means of click chemistry (e.g., azide-alkyne click chemistry) which facilitate visualization (e.g., fluorescent probes) as was demonstrated by R. Hatzenpichler et al. (Visualizing in situ translational activity for identifying and sorting slow-growing archaeal−bacterial consortia. PNAS, 2016).
The detailed characterization of active microorganisms at the single cell level and their identification can be performed by SERS. SERS is a method based on Raman microspectroscopy that uses metal nanoparticles (Ag or Au) to enhance the Raman signal. Therefore, SERS combines the ability of Raman microspectroscopy to deliver information about chemical structure at high spatial resolution with the advantage of specific visualization of cells in direct proximity to nanoparticles, and increases the signal by orders of magnitude up to 104 and higher. The analysis is conducted in a rapid and non-destructive way. Thus, BONCAT-SERS coupling has a potential to provide visualization and determination of chemical fingerprints of all active members (cells) of the community.
In the project, we plan to make screening of the suitable bacteria for model experiments, to prepare Ag and Au nanoparticles (NPs) for SERS analysis, to couple bacteria incorporating tagged AAAs with NPs via click reaction, to enable the detection and characterization of active microbial cells by combination BONCAT and SERS.
Publications
N. P. Ivleva, P. Kubryk & R. Niessner, Raman Microspectroscopy, Surface-enhanced Raman Scattering Microspectroscopy, and Stable-isotope Raman Microspectroscopy for Biofilm Characterization. Analytical & Bioanalytical Chemistry 2017, 409, 4253-4375 (invited review)
P. Kubryk, Niessner & N. P. Ivleva, On the Origin of the Band at around 730 cm-1 in SERS Spectra of Bacteria: Stable Isotope Approach. Analyst 2016, 141, 874-2878
P. Kubryk, J. Kölschbach, S. Marozava, T. Lüders, R. Meckenstock, R. Niessner & N. P. Ivleva; Exploring the Potential of Stable Isotope (Resonance) Raman Microspectroscopy and SERS for the Analysis of Microorganisms at Single Cell Level. Analytical Chemistry 2015, 87, 6622-6630